Category: Health Sciences and Medicine
ORIGINAL
Enhancing Deep Learning for Autism Spectrum Disorder Detection with Dual-Encoder GAN-based Augmentation of Electroencephalogram Data
Mejora del Aprendizaje Profundo para la Detección de Trastornos del Espectro Autista con Aumento de Datos de Electroencefalograma Basado en GAN de Codificador Dual
1Alliance College of Engineering and Design, Alliance University, Bangalore, India.
Cite as: Lalli K, M S. Enhancing Deep Learning for Autism Spectrum Disorder Detection with Dual-Encoder GAN-based Augmentation of Electroencephalogram Data. Salud, Ciencia y Tecnología - Serie de Conferencias. 2024; 3:958. https://doi.org/10.56294/sctconf2024958
Submitted: 13-02-2024 Revised: 03-05-2024 Accepted: 04-07-2024 Published: 05-07-2024
Editor: Dr.
William Castillo-González ![]()
ABSTRACT
Autism Spectrum Disorder (ASD) is a general neurodevelopmental condition that requires early and accurate diagnosis. Electroencephalography (EEG) signals are reliable biomarkers for ASD detection and diagnosis. A recent Deep Learning (DL) model called Resting-state EEG-based Hybrid Graph Convolutional Network (Rest-HGCN) has been developed for this purpose. However, a challenge in ASD diagnosis is the limited availability of EEG data, leading to imbalanced classes and ineffective model training. To address this issue, a new approach is proposed in this paper, which involves a generative model for EEG data augmentation. A novel Dual Encoder-Balanced Conditional Wasserstein Generative Adversarial Network (DEBCWGAN) is designed to produce fine synthetic minority-class EEG examples and augment the original training dataset. This model integrates the Variational Auto-Encoder (VAE) and balanced conditional Wasserstein GAN. Initially, EEG signals for ASD in the training dataset are pre-processed as Differential Entropy (DE) features and split into different segments. Each feature segment is processed in the temporal and the spatial domain depending on the electrode place. Then, twin encoders are trained to capture both spatial and temporal information from these features, concatenate them as Latent Variables (LVs), and provide them to the decoder to produce synthetic EEG examples. Additionally, gradient penalty and L2 regularization are used to speed up convergence and prevent overfitting effectively. Further, the augmented dataset is used to train the Rest-HGCN for ASD detection, enhancing its robustness and generalizability. Finally, test outcomes demonstrate that the DEBWGAN-GP-Rest-HGCN on the EEG Dataset for ASD and ABC-CT dataset achieves 91,6 % and 88,1 % accuracy, respectively compared to the Rest-HGCN, AlexNet, K-Nearest Neighbor (KNN) and Support Vector Machine (SVM).
Keywords: Autism Spectrum Disorder; EEG; Rest-HGCN; Class Imbalance; Data Augmentation; Differential Entropy; Dual-Encoder; Wasserstein GAN.
RESUMEN
El Trastorno del Espectro Autista (TEA) es una condición general del desarrollo neurológico que requiere un diagnóstico temprano y preciso. Las señales de electroencefalografía (EEG) son biomarcadores fiables para la detección y el diagnóstico de TEA. Para este propósito se ha desarrollado un modelo reciente de aprendizaje profundo (DL) llamado Red convolucional de gráficos híbridos basada en EEG en estado de reposo (Rest-HGCN). Sin embargo, un desafío en el diagnóstico de TEA es la disponibilidad limitada de datos de EEG, lo que genera clases desequilibradas y un entrenamiento de modelos ineficaz. Para abordar este problema, en este artículo se propone un nuevo enfoque, que implica un modelo generativo para el aumento de datos de EEG. Una novedosa Red Adversaria Generativa de Wasserstein Condicional Equilibrada con Codificador Dual (DEBCWGAN) está diseñada para producir excelentes ejemplos sintéticos de EEG de clase minoritaria y aumentar el conjunto de datos de entrenamiento original. Este modelo integra el codificador automático variacional (VAE) y la GAN Wasserstein condicional equilibrada. Inicialmente, las señales de EEG para TEA en el conjunto de datos de entrenamiento se preprocesan como características de entropía diferencial (DE) y se dividen en diferentes segmentos. Cada segmento de característica se procesa en el dominio temporal y espacial dependiendo de la ubicación del electrodo. Luego, se entrenan codificadores gemelos para capturar información espacial y temporal de estas características, concatenarlas como variables latentes (LV) y proporcionarlas al decodificador para producir ejemplos de EEG sintéticos. Además, la penalización de gradiente y la regularización L2 se utilizan para acelerar la convergencia y evitar el sobreajuste de forma eficaz. Además, el conjunto de datos aumentado se utiliza para entrenar Rest-HGCN para la detección de TEA, mejorando su solidez y generalización. Finalmente, los resultados de las pruebas demuestran que DEBWGAN-GP-Rest-HGCN en el conjunto de datos EEG para ASD y ABC-CT logra una precisión del 91,67 % y 88,1 %, respectivamente, en comparación con Rest-HGCN, AlexNet y K-Nearest Neighbor (KNN). y Máquina de vectores de soporte (SVM).
Palabras clave: Trastorno del Espectro Autista; EEG; Rest-HGCN; Desequilibrio de Clases; Aumento de Datos; Entropía Diferencial; Codificador Dual; Wasserstein GAN.
INTRODUCTION
ASD is a common developmental disorder that often emerges in infancy or early childhood. It is described by complications in public collaboration, restricted interests, monotonous activities, and sometimes intellectual disabilities.(1,2) A timely prognosis is vital for powerful intervention and support.(3) Behavioral and personality assessments are typically used to diagnose ASD, but the process might be laborious, tending to latencies in diagnosis.(4) Early identification of ASD is important as early intervention can help manage symptoms and bridge the gap between children with ASD and their typically developing peers.(5) Researchers are currently exploring the use of objective markers from Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) to discover ASD early.(6,7,8) However, diagnosing or screening ASD in children using these imaging techniques is not recommended due to the unclear correlation between neurological findings and ASD features. Previous studies have shown that children with autism exhibit less complex EEG signals as their brains develop, with distinct patterns in the right and middle brain regions related to typical kids.(9) This suggests that EEG signals can provide unique insights into the brain activity of autistic individuals. EEG signals offer complex temporal resolution, minor difficulty, and minimum expense compared to MRI and CT scans, making them a more practical diagnostic tool for ASD.(10) Additionally, EEG is utilized across varied ages and is more clinically accessible than MRI and CT. Hence, developing EEG-based models for autism diagnosis is essential for early detection and screening of ASD.
Researchers have investigated the application of DL techniques for ASD diagnosis using EEG data, citing their enhanced efficiency and cost-effectiveness.(11,12) Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) are frequently utilized DL models in ASD diagnosis.(13,27) Although CNN and RNN approaches have demonstrated some advancements in ASD diagnostics, there remains a necessity to fully utilize the graph information among EEG channels.(14) While CNNs excel at feature extraction from multi-channel data, they may not adequately capture the intricate relationships between channels, potentially affecting the prognostic precision of CNN- or RNN-based ASD frameworks.
As a result, Tang et al.(15) developed the Rest-HGCN model for ASD by combining the cognitive prior and the data-driven graph modules. The data-driven graph module captures information flow features dynamically, while the cognitive prior graph module utilizes the EEG brain functional links as prior graph data to learn robust functional connectivity configurations among brain areas. By leveraging attention mechanisms to obtain discriminative characteristics from both modules and sharing parameters, the Rest-HGCN enhances the distinction between healthy and ASD persons, leading to improved ASD performance.
Literature survey
This section discusses recent research on using EEG signals to classify children with autism. Kang et al.(16) applied the Minimum Redundancy Maximum Relevance (MRMR) attribute selection approach integrated with the SVM classifier to recognize children with ASD based on EEG and eye-tracking information. However, it included children aged 3 to 6, which limited the dataset size.
Oh et al.(17) developed a classification system to distinguish between ASD and healthy EEG waves. They extracted nonlinear characteristics from the brain signs, selected the most relevant characteristic using a t-test, and reduced them using Marginal Fisher Analysis (MFA). The selected features were ranked based on their t values and input into SVM models with polynomial degrees 1, 2, and 3. Additionally, 3 most relevant characteristics were utilized to create an index for ASD diagnosis. Conversely, manual feature extraction can be labor-intensive.
Alturki et al.(18) developed a diagnostic system for epilepsy and ASDs using EEG signals. It utilized Independent Component Analysis (ICA) to remove artifacts, followed by segmentation and noise removal using a band-pass filter. Features were extracted using a Common Spatial Pattern (CSP), including variance, entropy, energy, and Local Binary Pattern (LBP). Classification was performed using Linear Discriminant Analysis (LDA), SVM, KNN and Artificial Neural Network (ANN). However, it was limited by the dataset used.
Baygin et al.(19) developed a hybrid lightweight deep feature extractor for a large EEG dataset of autism patients and controls. They introduced a signal-to-image conversion model, extracting features from EEG signals with 1D-LBP and Short Time Fourier Transform (STFT) to create spectrogram images. Then, deep features were extracted using pre-trained MobileNetV2, ShuffleNet, and SqueezeNet models. Feature ranking and selection were done with a two-layered ReliefF algorithm, followed by the SVM classification. However, it was limited by a small dataset.
Tawhid et al.(20) developed a diagnostic framework for identifying ASD using time-frequency spectrogram images of EEG signals. The raw EEG signals were pre-processed and transformed into 2D spectrogram images using STFT. Machine learning and DL techniques were then utilized to analyze the images. In the machine learning approach, textural features were extracted and fed into six machine learning classifiers. In the DL approach, CNN model was tested. However, the study was limited by a small dataset of 16 subjects (12 ASD, 4 control), which may affect the generalizability of the results.
Sinha et al.(21) proposed an effective method for autism detection using EEG signals. They preprocessed pre-recorded EEG signals with a digital filter and extracted features in the time and frequency domains using discrete wavelet transform. These features were then inputted into various classifiers, including neural networks, SVM, KNN, subspace KNN, and LDA, for ASD detection. However, the limited availability of autistic EEG datasets restricted the model accuracy to some extent.
Liao et al.(23) introduced a machine learning method that combines EEG, eye fixation, and facial expression data to identify children with ASD. They developed a novel feature extraction method for these modalities and used a weighted naive Bayes algorithm for multimodal fusion, achieving an accuracy of 87,50 % in classification. However, the study had a limited sample size. Menaka et al.(24) developed an enhanced AlexNet with Linear Frequency Cepstral Coefficients (LFCC) model to classify ASD using EEG signals. The model achieved 90 % accuracy, but its drawback was the reliance on small sample datasets, which hindered the validation of the model’s robustness due to the lack of larger datasets. Earlier research on using EEG signals for classifying ASD faces challenges such as small dataset sizes, lack of data from younger patients, and limited availability of autistic EEG datasets. To address these gaps, this study aims to create larger and more diverse EEG datasets, including data from younger patients with ASD using a novel deep generative network, to enhance the accuracy and robustness of ASD classification systems based on EEG signal data.
METHOD
This section offers a comprehensive explanation of the DEBCWGAN model. Figure 1 depicts the study’s conceptual diagram, outlining the various stages: EEG dataset acquisition, preprocessing, EEG augmentation, training, and classification.
EEG Dataset Details
This work uses two well-known datasets to collect EEG signals from children with ASD and Typically Developing (TD) children:
1. EEG Dataset for ASD:(25,26) in this dataset, EEG data were collected using the Biosemi Active two EEG system and converted to .set and .fdt files via EEGLAB. Each recording includes a .fdt file with the data and a .set file with recording parameters. The dataset consists of recordings from 28 persons with ASD and 28 with TD aged 18-68. The recordings were taken throughout a 2,5-minute (150-second) period of eyes closed and sleeping. These files can be viewed using EEGLAB software. For training, 44 data samples (22 from each class) are used, while the remaining 12 data samples (6 from each class) are used for testing. In this study, the DEBCWGAN model generates 2 000 artificial EEG data (1 000 from each class) using 44 training samples. These generated data are combined with the original training samples to improve the classification performance.
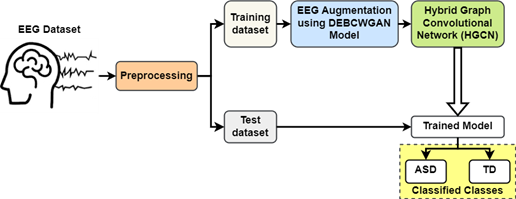
Figure 1. Conceptual Design of the Presented Work
1. Autism Biomarkers Consortium for Clinical Trials (ABC-CT) dataset:(22,28) this dataset includes EEG and eye-tracking data from 280 contributors with ASD and 119 with TD, aged 6–11 years, from five sites. Participants had IQs ranging from 60–150 for ASD and 80–150 for TD. EEG data was collected at three-time points: baseline, 6 weeks later, and 6 months later. 211 ASD and 106 TD contributors offered EEG during the first-time step, and 215 ASD and 100 TD contributors during the 2nd time step. EEG signals were gathered using 128-channel equipment at a sampling frequency of 1 000 Hz. For training, 319 samples (224 from ASD and 95 from TD) are used, while the remaining 84 samples (60 from ASD and 24 from TD) are used for testing. In this study, the DEBCWGAN model generates 2 000 artificial EEG data (1 000 from each class) using 319 training samples. These generated data are combined with the original training samples to improve the classification performance. Figure 2 shows raw EEG waves from both healthy and ASD individuals.
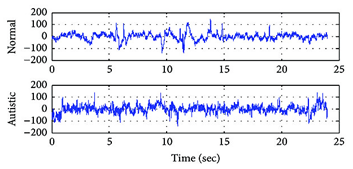
Figure 2. Illustration of Original Healthy and ASD EEG Signals
Preprocessing
The raw EEG signal contains both relevant and redundant information. Directly generating the raw signal using a generator (G) may be affected by this redundancy. Thus, capturing EEG characteristics will circumvent interloping from unwanted data and provide more specific information for ASD detection. This study enhances the dataset by generating artificial samples of DE. DE is a feature extracted from EEG signals in 5 spectrums (Delta δ: 1-3Hz, Theta θ: 4-7Hz, Alpha α: 8-13Hz, Beta β: 14-30Hz, Gamma γ: 31-50Hz). Figure 3 compares the five different EEG bands.(29,30)
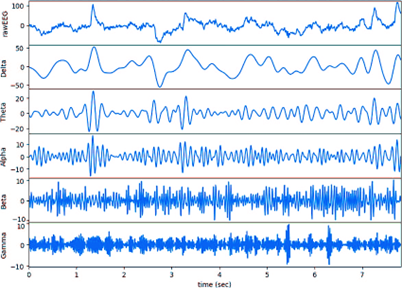
Figure 3. Comparison of EEG Bands
The DE value is the logarithmic spectral power of the EEG pattern in a specific spectrum. The DE calculation for a frequency band is given in equation 1, where Pi represents the energy spectrum corresponding to the wave disparity multiplied by a fixed coefficient N, which is the predetermined time window length.
![]()
As the DE feature does not include information about electrode position distribution and timing, the processed DE characteristic was utilized as the network input. Each unit period’s DE feature obtained from the SEED database has a volume of 310, with sizes of 62 channels and 5 spectrums. Traditional GAN models focus on features within specific periods, neglecting time and spatial relationships across constant intervals. This study introduces a process to overcome this restriction. Figure 4 shows the process of reducing the DE of the EEG for a specific person. Initially, t seconds of DE characteristics are extracted constantly from the initial to t seconds. To maintain data reliability during learning, when the subsequent characteristics is lower than t seconds, the preceding characteristics is utilized to compensate. In this study, t is set to 5 seconds. If the duration of data for a person in a test is T seconds, then T segments of characteristics in t seconds are acquired.
A slice of data consists of 5 seconds of characteristics across 5 spectrums (b) and 62 channels (c). This data is then transformed into a 5-second segment with 310 dimensions, focusing solely on timing information without channel position details. The transformed data is represented as 5 seconds across 5 bands with a grid size of 16×16. This transformation involves projecting the original 62-channel data onto a 2D grid using polar coordinates and interpolating using the Clough-Tocher method. The final feature format is 25×16×16, capturing relative electrode positions without time sequence information.
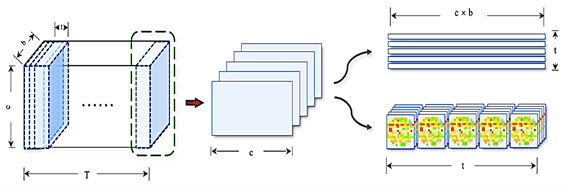
Figure 4. Process of Reducing Differential Entropy Features
DEBCWGAN Model for EEG Data Augmentation
The VAE-GAN model combines the discriminator (D)’s analysis of EEG signal properties with the VAE’s reconstruction error.(25) This integration includes the feature encoding of actual data in the GAN. The VAE’s decoder is referred to as G. This study uses two encoders with GAN to learn spatiotemporal information from EEG signals. The architecture of proposed DEBCWGAN model is portrayed in figure 5. First, Encoder-I extracts time sequence information, while Encoder-II extracts spatial distribution information. Then, LV distributions ztime and zspace are sampled from the encoders, and concatenated to form a new distribution zconcat.
So, the loss function of the Dual Encoder GAN (DEGAN) model is formulated as:
![]()
In equation 2, the initial expression represents the KL loss in VAE, the 2nd expression represents the restoration loss. In the D’s initial layer, the information sampled from a combination of the produced and real data. Also, the 3rd expression represents the D loss.

Figure 5. Structure of DEBCWGAN Model for EEG Data Augmentation
To stabilize GAN training, this study incorporates the balanced conditional Wasserstein GAN principle into DEGAN, resulting in a new loss function of the DEBCWGAN model is as follows:
![]()
On the other hand, the use of weight clipping in DEBCWGAN can lead to slow convergence. To address this, a gradient penalty term is added to the input instead of weight clipping. This modification aims to improve the performance of DEBCWGAN. The loss function is as follows:
![]()
In equations 4, 5, 6, ‖∙‖2 is the 2-norm, x̂ is attained by random interpolation sampling on the line between the actual example x and the produced example G(zconcat│y), LGP is the loss function of D in DEBCWGAN, while adding the GP term to its input. Also, x̂ is calculated by:
![]()
In equation 5, ε follows a rectangular distribution on [0,1]. Additionally, the L2 regularization is added to facilitate G to produce information analogous to the actual data and avoid overfitting. So, the loss function is as follows:
![]()
In equation 6, L(L2) is the loss function of G, while adding the standard loss L2 distance into DEBCWGAN. At last, the loss function is a mixture of DEBCWGAN’s actual loss function, GP term and L2-norm.
![]()
In equation 7, λ1 and λ2 are the coefficients of GP and L2 regularization, respectively. In the learning stage, D and G engage in discontinuous opposition learning. The Adam optimizer is applied to fine-tune the parameters of the whole network.
Rest-HGCN for ASD Detection
The artificial EEG data is merged with real data to create a new training dataset. This dataset is then used to train the Rest-HGCN classifier, achieving high accuracy in classifying children with ASD and normal children. The training process of DEBCWGAN+Rest-HGCN is illustrated in figure 6.
Algorithm 1: DEBCWGAN+Rest-HGCN Model for ASD Classification
Input: real EEG data xr
Output: generated data xg
Begin
1. Initialize DEBCWGAN+Rest-HGCN parameters;
2. while(epoch<Maximum number of epochs)
3. Processing the EEG data into two forms (temporal and spatial features);
4. Calculate =E1(xr ), zspace=E2(xr);
5. Concatenate ztime and zspace into zconcat;
6. Obtain the generated data via the decoder;
7. Determine D’s loss and modify the network;
8. Determine the loss of DEBCWGAN and modify the decoder unit;
9. Determine G’s loss and modify G unit;
10. epoch=epoch+1;
11. End while
12. Combine the generated artificial samples with the real training data to create an augmented dataset.
13. Train the Rest-HGCN classifier using the augmented EEG dataset;
14. Capture dynamic information flow from the cognitive prior and data-driven graph modules in the Rest-HGCN;
15. Fuse discriminative characteristics from both modules using attention;
16. Test the trained Rest-HGCN model using test EEG dataset to classify children with ASD and TD;
17. End
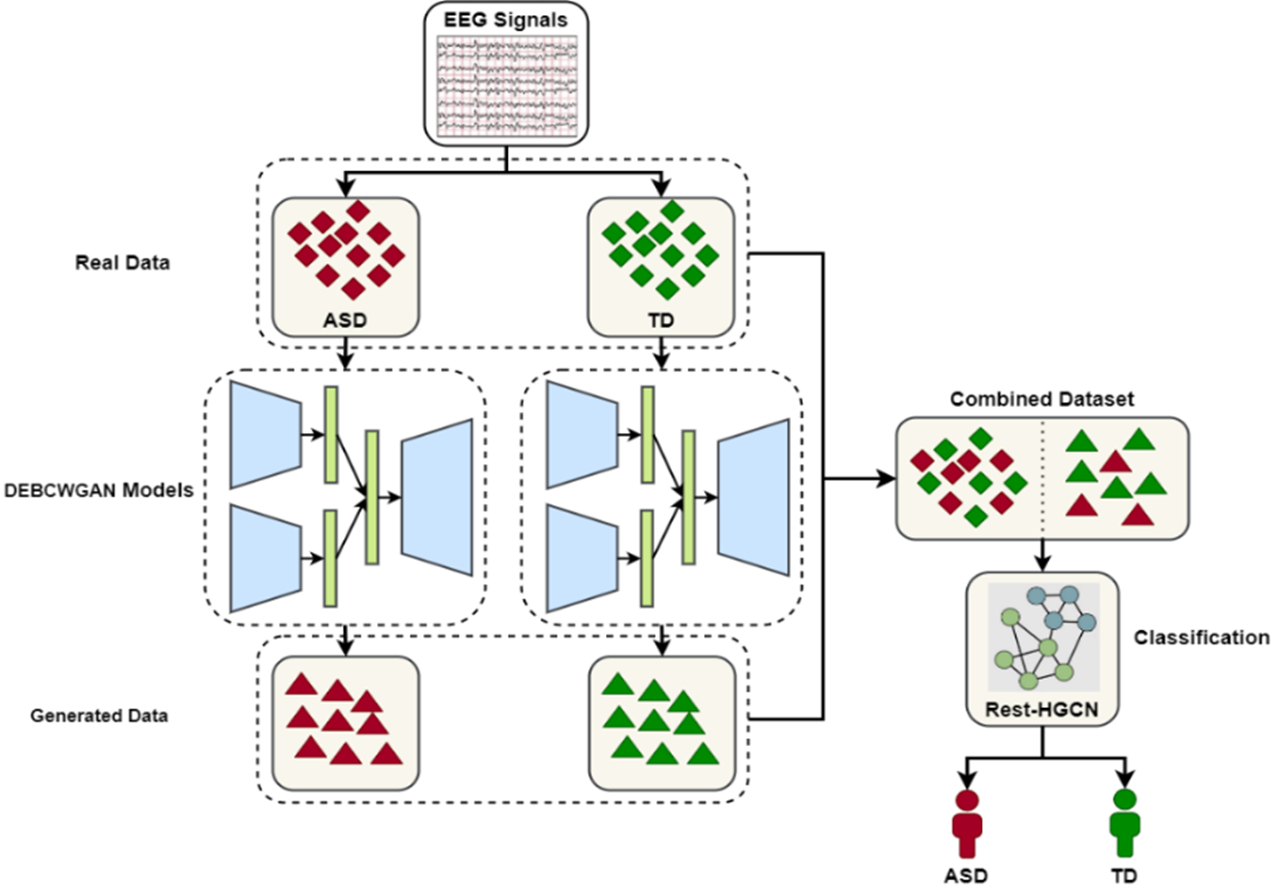
Figure 6. Flow Diagram of EEG Data Augmentation and ASD Classification Based on DEBCWGAN+Rest-HGCN Model
RESULTS
This section evaluates the robustness of DEBCWGAN+Rest-HGCN model on two different EEG datasets described in Section 3.1. The performance is compared with the existing models such as Rest-HGCN(14), SVM(15), CSP-LBP-KNN(17) and AlexNet-LFCC.(22) The tests are conducted on a laptop equipped with an Intel® Core TM i5-4210 CPU @ 3GHz, 4GB RAM, and a 1TB HDD running Windows 10 64-bit. To evaluate the performance improvements, all existing and proposed models are implemented in the MATLAB 2019b software with two different datasets.
Parameter Settings
Table 1 provides a list of parameters used to simulate both the existing and proposed models for performance evaluation.
|
Table 1. List of Parameters for Proposed and Existing Models |
|
|
Parameters |
Range |
|
Proposed DEBCWGAN+Rest-HGCN, Rest-HGCN(14), SVM(15), CSP-LBP-KNN(17) and AlexNet-LFCC(22) |
|
|
Learning rate |
0,001 |
|
Dropout rate |
0,5 |
|
Epochs |
120 |
|
Batch size |
32 |
|
Optimizer |
Adam |
|
Loss function |
Cross-entropy |
|
SVM(15) |
|
|
Kernel category |
Linear |
|
Kernel degree |
2 |
|
Penalty |
0,1 |
|
Y |
0,01 |
|
CSP-LBP-KNN(17) |
|
|
No. of neighbors, k |
12 |
|
Distance metric |
Euclidean |
|
AlexNet-LFCC(22) |
|
|
No. of convolution layer |
5 |
|
No. of pooling layer |
2 |
|
Depth |
96 |
|
Filter size |
3,3 |
|
Classifier |
Softmax |
|
Activation function |
ReLU |
|
Rest-HGCN(14) |
|
|
No. of graph convolution layer |
3 |
|
No. of hidden layer |
1 |
|
No. of neurons in each hidden layer |
15 |
|
Proposed DEBCWGAN+Rest-HGCN |
|
|
No. Of G |
1 |
|
No. Of D |
1 |
|
Activation function |
LeakyReLU |
|
λ1 |
10 |
|
λ2 |
100 |
Performance Evaluation Metrics
The model’s ability to detect ASD is evaluated using the following performance metrics:
· Accuracy is the percentage of correctly detected samples out of the total samples tested.
![]()
In equation 8, TP defines the situation where the network accurately categorized that a subject has ASD when they have ASD. TN represents the situation where the network accurately categorized that a subject does not have ASD when they truly do not have ASD. FP denotes the situations where the network inaccurately categorized that a subject has ASD when they do not have ASD. FN indicates the situation where the network inaccurately categorized that a subject does not have ASD when they do have ASD.
· Precision: it is computed as:
![]()
· Recall: it is measured by
![]()
· F1-score: it is determined by
![]()
A study was conducted on the confusion matrix to evaluate the DEBCWGAN+Rest-HGCN efficiency in distinguishing between healthy and ASD patients. The outcomes are shown in figure 7, highlighting the classification accuracy.
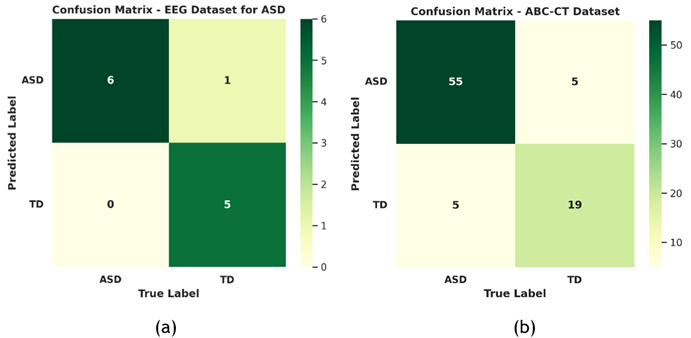
Figure 7. Confusion Matrix of DEBCWGAN+Rest-HGCN Model. (a) EEG Dataset for ASD and (b) ABC-CT Dataset
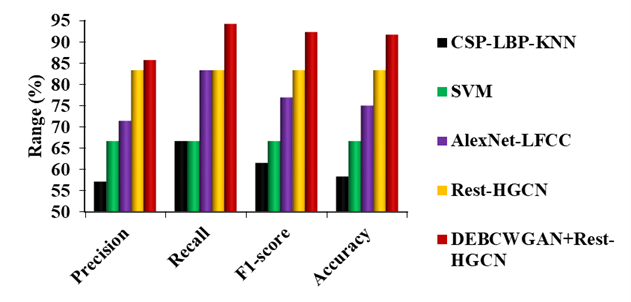
Figure 8. Comparison of DEBCWGAN+Rest-HGCN Model against Existing Models on EEG Dataset for ASD
Figure 8 shows the comparison of proposed and existing models on the EEG Dataset for ASD. It is observed that the DEBCWGAN+Rest-HGCN outperforms other models in classifying children with ASD due to effective model training using large-scale training dataset. Compared to CSP-LBP-KNN, SVM, AlexNet-LFCC and Rest-HGCN, DEBCWGAN+Rest-HGCN increases the precision by 50 %, 28,56 %, 19,99 %, and 2,86 %, recall by 41,32 %, 41,32 %, 13,07 %, and 13,07 %, F1-score by 50 %, 38,46 %, 20,01 %, and 10,78 %, and accuracy by 57,16 %, 37,5 %, 22,23 %, and 10,01 %, respectively.
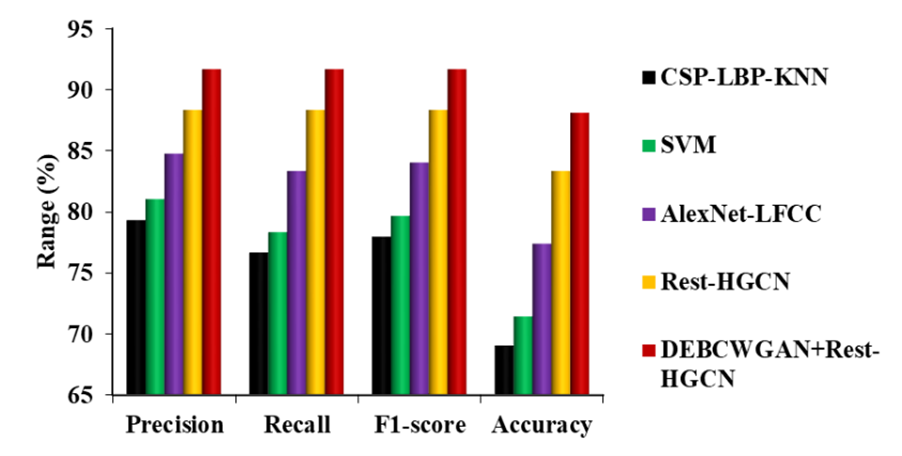
Figure 9. Comparison of DEBCWGAN+Rest-HGCN Model against Existing Models on ABC-CT Dataset
Figure 9 shows the comparison of proposed and existing models on the ABC-CT dataset. It is observed that the DEBCWGAN+Rest-HGCN outperforms other models in classifying children with ASD due to effective model training using large-scale training dataset. Compared to CSP-LBP-KNN, SVM, AlexNet-LFCC and Rest-HGCN models, DEBCWGAN+Rest-HGCN achieves higher precision, recall, F1-score, and accuracy. Precision is improved by 15,58 %, 13,13 %, 8,17 %, and 3,78 %, recall by 19,56 %, 17,03 %, 10,01 %, and 3,78 %, F1-score by 17,57 %, 15,08 %, 9,09 %, and 3,78 %, and accuracy by 27,59 %, 23,34 %, 13,85 %, and 5,72 %, respectively.
CONCLUSIONS
This study proposed a novel DEBCWGAN model combined with the Rest-HGCN classifier for accurate detection of ASD using EEG signals. The DEBCWGAN incorporates dual encoders to extract both temporal and spatial information from DE features of the EEG. This allows it to produce fine synthetic minority-class examples to augment the original imbalanced training dataset. The augmented dataset is then used to train the Rest-HGCN model, enhancing its robustness and generalization ability. Experimental results on two public EEG datasets demonstrate that the DEBCWGAN+Rest-HGCN model outperforms existing techniques like SVM, KNN, AlexNet, and the original Rest-HGCN in accurately classifying children with ASD versus neurotypical controls. The proposed DEBCWGAN+Rest-HGCN model achieves classification accuracies of 91,67 % on the EEG Dataset for ASD and 88,1 % on the ABC-CT dataset, marking a significant improvement over prior method. This study highlights the promise of DL and data augmentation schemes for advancing the early recognition and diagnosis of ASD based on EEG biomarkers.
BIBLIOGRAPHIC REFERENCES
1. Hadders‐Algra M. Emerging signs of autism spectrum disorder in infancy: Putative neural substrate. Dev Med Child Neurol. 2022;64(11):1344-50.
2. Riglin L, Wootton RE, Thapar AK, Livingston LA, Langley K, Collishaw S, et al. Variable emergence of autism spectrum disorder symptoms from childhood to early adulthood. Am J Psychiatry. 2021;178(8):752-60.
3. McCarty P, Frye RE. Early detection and diagnosis of autism spectrum disorder: Why is it so difficult? Semin Pediatr Neurol. 2020;35:100831.
4. Kaufman NK. Rethinking “gold standards” and “best practices” in the assessment of autism. Appl Neuropsychol Child. 2022;11(3):529-40.
5. Iadarola S, Pellecchia M, Stahmer A, Lee HS, Hauptman L, Hassrick EM, et al. Mind the gap: An intervention to support caregivers with a new autism spectrum disorder diagnosis is feasible and acceptable. Pilot Feasibility Stud. 2020;6:1-13.
6. Lalli, K., & Senbagavalli, M. (2023). Identification of Biomarker for Autism Spectrum Disorder Using Eeg: A Review. Proceedings-2023 International Conference on Advanced Computing and Communication Technologies, ICACCTech 2023.
7. Haweel R, AbdElSabour Seada N, Ghoniemy S, ElBaz A. A review on autism spectrum disorder diagnosis using task-based functional MRI. Int J Intell Comput Inf Sci. 2021;21(2):23-40.
8. Moridian P, Ghassemi N, Jafari M, Salloum-Asfar S, Sadeghi D, Khodatars M, et al. Automatic autism spectrum disorder detection using artificial intelligence methods with MRI neuroimaging: A review. Front Mol Neurosci. 2022;15:999605.
9. Kurkin S, Smirnov N, Pitsik E, Kabir MS, Martynova O, Sysoeva O, et al. Features of the resting-state functional brain network of children with autism spectrum disorder: EEG source-level analysis. Eur Phys J Spec Top. 2023;232(5):683-93.
10. Stoyell SM, Wilmskoetter J, Dobrota MA, Chinappen DM, Bonilha L, Mintz M, et al. High-density EEG in current clinical practice and opportunities for the future. J Clin Neurophysiol. 2021;38(2):112-23.
11. Kohli M, Kar AK, Sinha S. The role of intelligent technologies in early detection of autism spectrum disorder (ASD): A scoping review. IEEE Access. 2022;10:104887-913.
12. Jui SJJ, Deo RC, Barua PD, Devi A, Soar J, Acharya UR. Application of entropy for automated detection of neurological disorders with electroencephalogram signals: A review of the last decade (2012-2022). IEEE Access. 2023;11:71905-24.
13. Khodatars M, Shoeibi A, Sadeghi D, Ghaasemi N, Jafari M, Moridian P, et al. Deep learning for neuroimaging-based diagnosis and rehabilitation of autism spectrum disorder: A review. Comput Biol Med. 2021;139:104949.
14. Ahmedt-Aristizabal D, Armin MA, Denman S, Fookes C, Petersson L. Graph-based deep learning for medical diagnosis and analysis: Past, present and future. Sensors. 2021;21(14):4758.
15. Tang T, Li C, Zhang S, Chen Z, Yang L, Mu Y, et al. A hybrid graph network model for ASD diagnosis based on resting-state EEG signals. Brain Res Bull. 2024;206:110826.
16. Kang J, Han X, Song J, Niu Z, Li X. The identification of children with autism spectrum disorder by SVM approach on EEG and eye-tracking data. Comput Biol Med. 2020;120:103722.
17. Oh SL, Jahmunah V, Arunkumar N, Abdulhay EW, Gururajan R, Adib N, et al. A novel automated autism spectrum disorder detection system. Complex Intell Syst. 2021;7(5):2399-413.
18. Alturki FA, Aljalal M, Abdurraqeeb AM, Alsharabi K, Al-Shamma’a AA. Common spatial pattern technique with EEG signals for diagnosis of autism and epilepsy disorders. IEEE Access. 2021;9:24334-49.
19. Baygin M, Dogan S, Tuncer T, Barua PD, Faust O, Arunkumar N, et al. Automated ASD detection using hybrid deep lightweight features extracted from EEG signals. Comput Biol Med. 2021;134:104548.
20. Tawhid MNA, Siuly S, Wang H, Whittaker F, Wang K, Zhang Y. A spectrogram image based intelligent technique for automatic detection of autism spectrum disorder from EEG. PLoS One. 2021;16(6).
21. Chavez-Cano AM. Artificial Intelligence Applied to Telemedicine: opportunities for healthcare delivery in rural areas. LatIA 2023;1:3-3. https://doi.org/10.62486/latia20233.
22. Sinha T, Munot MV, Sreemathy R. An efficient approach for detection of autism spectrum disorder using electroencephalography signal. IETE J Res. 2022;68(2):824-32.
23. Lalli, K., Shrivastava, V. K., & Shekhar, R. (2023, April). Detecting Copy Move Image Forgery using a Deep Learning Model: A Review. In 2023 International Conference on Artificial Intelligence and Applications (ICAIA) Alliance Technology Conference (ATCON-1) (pp. 1-7). IEEE.
24. Liao M, Duan H, Wang G. Application of machine learning techniques to detect the children with autism spectrum disorder. J Healthc Eng. 2022;2022:1-10.
25. Menaka R, Karthik R, Saranya S, Niranjan M, Kabilan S. An improved AlexNet model and cepstral coefficient-based classification of autism using EEG. Clin EEG Neurosci. 2024;55(1):43-51.
26. Dickinson A, Jeste S, Milne E. Electrophysiological signatures of brain aging in autism spectrum disorder. Cortex. 2022;148:139-51.
27. McPartland JC, Bernier RA, Jeste SS, Dawson G, Nelson CA, Chawarska K, et al. The autism biomarkers consortium for clinical trials (ABC-CT): Scientific context, study design, and progress toward biomarker qualification. Front Integr Neurosci. 2020;14:16.
28. Suárez YS, Laguardia NS. Trends in research on the implementation of artificial intelligence in supply chain management. LatIA 2023;1:6-6. https://doi.org/10.62486/latia20236.
29. Senbagavalli, M. (2022). Artificial Intelligence and Medical Information Modeling. In Handbook of Research on Mathematical Modeling for Smart Healthcare Systems (pp. 1-11). IGI Global.
30. Larsen ABL, Sønderby SK, Larochelle H, Winther O. Autoencoding beyond pixels using a learned similarity metric. In: International Conference on Machine Learning; 2016. p. 1558-66.
FINANCING
No financing.
CONFLICT OF INTEREST
None.
AUTHORSHIP CONTRIBUTION
Conceptualization: K Lalli, Senbagavalli M.
Data curation: K Lalli, Senbagavalli M.
Formal analysis: K Lalli, Senbagavalli M.
Research: K Lalli, Senbagavalli M.
Methodology: K Lalli, Senbagavalli M.
Resources: K Lalli, Senbagavalli M.
Software: K Lalli, Senbagavalli M.
Validation: K Lalli, Senbagavalli M.
Drafting - original draft: K Lalli, Senbagavalli M.
Writing - proofreading and editing: K Lalli, Senbagavalli M.